Surrey AKI Detection Algorithm (SAKIDA) - User Manual
About SAKIDA
This webpage is set up to provide some support for you to use the Surrey Acute Kidney Injury Detection Software (SAKIDS) which is based on the SAKIDA algorithm. You should read the SAKIDA software introduction before continuing to read this page which serves as the user manual. To download the SAKIDA software, visit this page.
1. Get familiar with the interface
First, get to know the way your interact with the software in five steps. Don’t forget to press F1 any time to get to this help website.
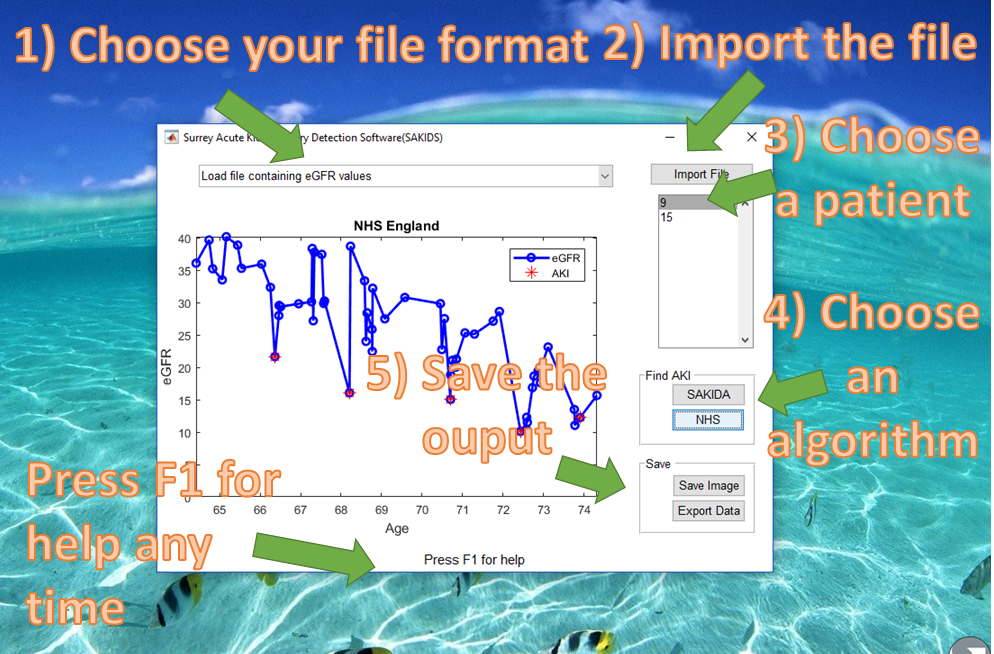
1) Choose your file format
There are four file formats. Please choose the right option. If you are not sure, please choose the first option, which loads serum creatinine in mg/dL and then computes eGFR using the CKD-EPI equation as our software can figure out automatically the correct formula to used. If you want to use the MDRD equation, then you should choose the third option.

Below, we shall show two cases: files with creatinine as input or eGFR as input
Input file with Creatinine measurements
Let’s take a look at an example file below: Example_Scr_in_micromolperL3.csv.
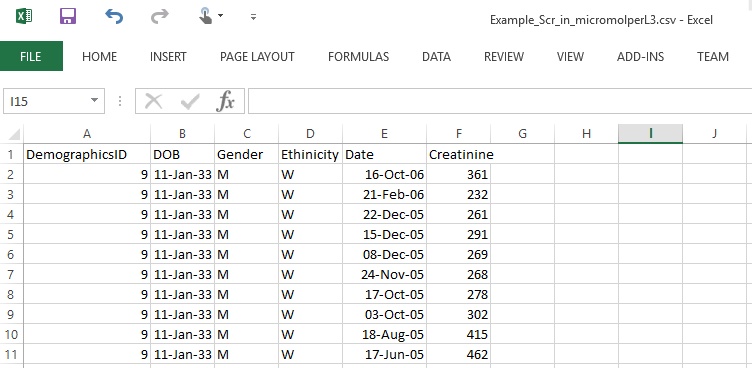
All the headings above mandatory; otherwise, the loading will fail.
- DemographicsID - a unique patient ID (integer)
- DOB. This is a date of the format dd-mmm-yy, e.g., 11-Jan-33, which is taken as 11 Jan 1933. The software can also accept the American date format: Jan-11-33. If 11-01-33 is presented, the software will interpret this as 1 November 1933, which is wrong. If your date is in dd-mm-yy or mm-dd-yy (e.g., 11-01-33 or 01-11-33), which can create a lot of confusion, we recommend that you change your format automatically using Excel, following the instructions here.
- Gender: should be M or F
- Ethnicity can be any character, e.g., ‘W’ for white, but our software will only look for the flag ‘B’ for black.
- Date: This is the date the measurement was taken.
- Creatinine: The serum creatinine value in micro m/L or mg/dL. To distinguish the two, choose the right option as stated above.
All fields can be named exactly as they are: DemographicsID (and not ID, please); DOB (and not Date of Birth); etc. Our software can only recognise these headings.
Input file with eGFR
An example file with eGFR is also provided: Example_eGFR.csv.
If your file has pre-computed eGFR values, then you should have Date, Age, and eGFR fields (attributes).
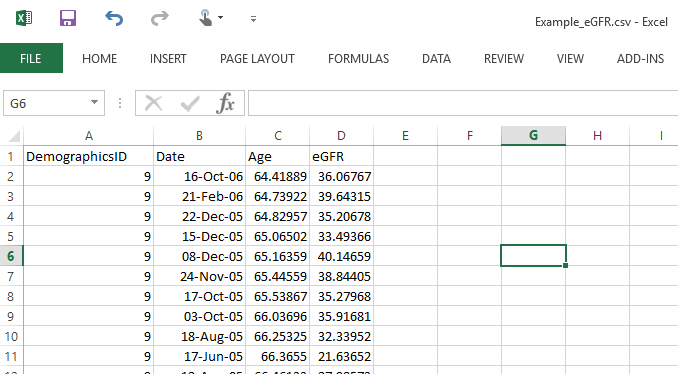
Note that we don’t need the gender, ethnicity fields, etc.
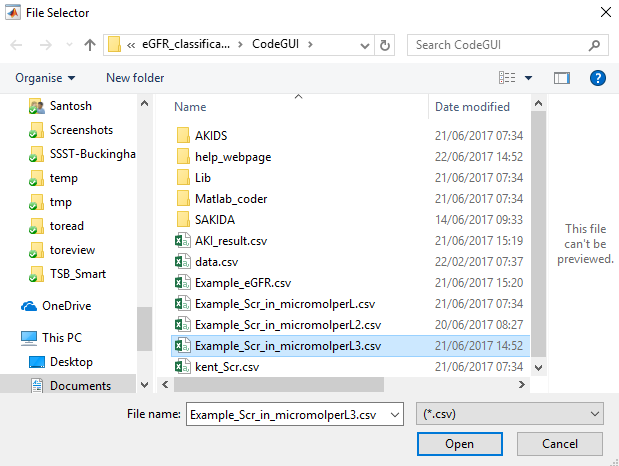
2) Import the file
Next, import the file.
3) Choose a patient
4) Choose an AKI detection algorithm
5) Save the output
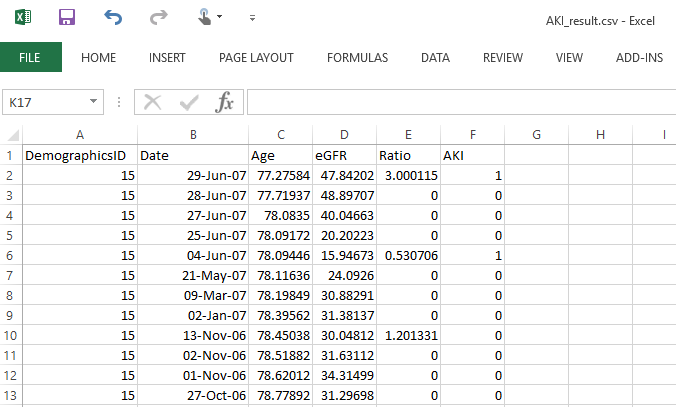
If you have chosen to save an image, the filename is AKI.png which is stored in the same directory as where the software is installed. The exported data is saved in a default file named ‘AKI_result.csv’. An example of output is shown below. Note that we calculate the age by assuming one year has 365.25 days.
What’s next?
If you haven’t downloaded the SAKIDA software, do this here.
If you want to use the software for your research or to run it on a large database of patients, please do get in touch with me. I will be thrilled to hear from you.
Cite this blog post
Bibtex
@misc{ poh_2018_01_14_sakida-help,
author = {Norman Poh},
title = { Surrey AKI Detection Algorithm (SAKIDA) - User Manual },
howpublished = {\url{ http://normanpoh.github.com/blog/2018/01/14/sakida-help.html},
note = "Accessed: ___TODAY___"
}